Two out of the three samples I sent out for sequencing came back with results. My first unknown sample sample was a bit noisy, so I only blasted 687 nucleotides. The closest genetic match was Acinectobacter baylyi strain B2 (Query 100%, Identity 96%). The majority of the bases that did not match were “n” so they could have been any nucleotide. Ironically, this is one of the tester strains used in lab. Colonies are described as “circular, convex, smooth and slightly opaque,” and the genus has general characteristics such as being aerobic, gram-negative bacilli (Carr et. al.). This corresponds with my data. I put the picture of my gram stain below, because initially I thought that I had spore formation or a mix of 2 colonies, but with the BLAST data I can probably say this is not spore formation and is indeed gram-negative.
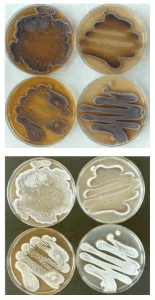
For my second sample, I was able to run 792 bases. The BLAST results only had members of the genus streptomyces. The closest match was Streptomyces spororaveus strain NBRC 15456 (query 99%, identity 99%). Streptomyces spororaveus gram positive, forms dusty gray multicellular complexes, as well as spore chains, which I observed in lab (Podstawka). The spore chains made it hard to pick, spread, and gram stain this organism. Below is an image of the spore chains from Streptomyces spororaveus, as well as spread plates on various media. My spread plates looked very similar.
Image citation: Joachim M. Wink, HZI – Helmholtz-Zentrum für Infektionsforschung GmbH, Inhoffenstr. 7, 38124 Braunschweig, Germany
Streptomyces is in the domain Bacteria, phylum Actinobacteria, class Actinobacteria, order Actinomycetales, and family Streptomycetaceae (Podstawka). Optimal growth occurs at 28 degrees celsius, and streptomyces are commonly found in soil. (Podstawka). Streptomyces can survive in vastly diverse environmental / nutritional conditions due to their ability to form spores, and commonly form symbiotic relationships with plant roots (de Lima Procópio). “The most interesting property of Streptomyces is the ability to produce bioactive secondary metabolites, such as antifungals, antivirals, antitumorals, anti-hypertensives, immunosuppressants, and especially anitbiotics” (de Lima Procópio). Over 60% of naturally derived antibiotics were isolated from members of the genus streptomyces. The first, streptothricin, was discovered in 1942 (Wavte). It is predicted that members of the genus streptomyces are capable of producing over 100,000 secondary metabolites that have antibiotic properties (Wavte). The antibiotic Streptomicin is derived from S. griseus, and Avermictin is derived from S. avermitilis (de Lima Procópio). Notice below how many antibiotics are from S. ___ indicating they are derived from streptomyces.
Image source: de Lima Procópio, Rudi Emerson. “Antibiotics produced by Streptomyces.” Braz journal of infect dis. 2012;16(5):466–471
Carr, E. L. “Seven Novel Species of Acinetobacter Isolated from Activated Sludge.” International Journal Of Systematic And Evolutionary Microbiology, vol. 53, no. 4, 2003, pp. 953–963., doi:10.1099/ijs.0.02486-0.
de Lima Procópio, Rudi Emerson. “Antibiotics produced by Streptomyces.” Braz journal of infect dis. 2012;16(5):466–471
Podstawka, Adam. “Streptomyces Spororaveus | Type Strain | DSM 41462, ATCC 43694, INMI 101, VKM Ac-318 | BacDiveID:16196.” BacDive | The Bacterial Diversity Metadatabase, bacdive.dsmz.de/strain/16196.
Watve, Milind, et al. “How Many Antibiotics Are Produced by the Genus Streptomyces ?” Archives of Microbiology, vol. 176, no. 5, 2001, pp. 386–390.,