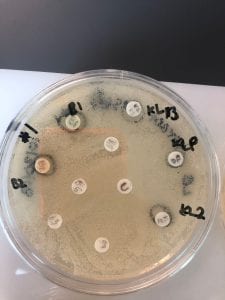
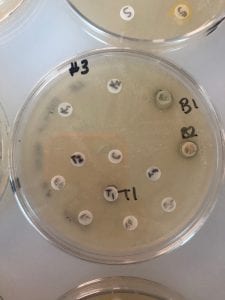
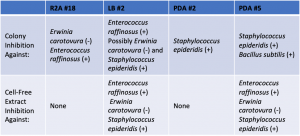
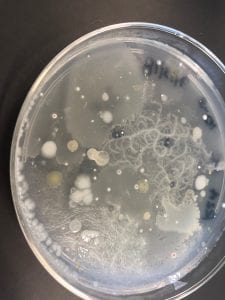
- Assigned ESKAPE Pathogen
Staphylococcus Aureus
- Why is this ESKAPE Pathogen of interest (in brief)
Staphylococcus Aureus(S. aureus)is a fairly common bacteria, anywhere from 30 to 50 percent of humans have this bacteria growing on them (1, 2). Typically, S. aureus is found on human skin and mucous membranes, and is not of any concern until it crosses into the bloodstream or internal tissues (2). Infections typically are spread in clinical and hospital settings, but can also occur in non-clinical environments (2). The major concern regarding S. aureusis a particular strain, methicillin-resistantS. aureus, or MRSA, which is a highly difficult form of S. aureus to treat due to resistance and has a very high morbidity rate (3).
- General Cellular and Morphological Characteristics of the Organism (taxonomic classification, nutrition, cell shape, habitat)
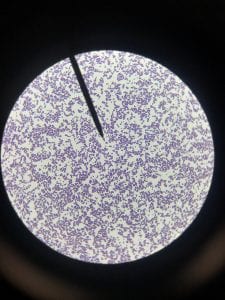
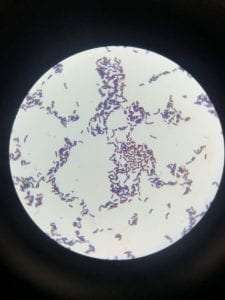
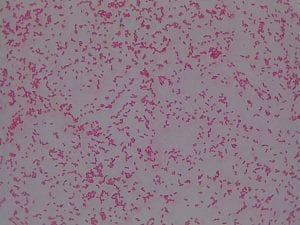
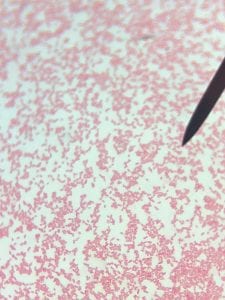
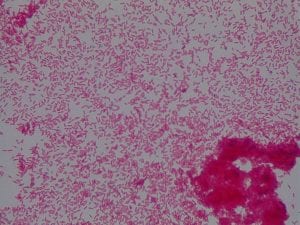
Staphylococcus aureusis a Gram+bacteria with a cocci shape (round, spherical shape) and is a member of the Staphylococcaceae family (2). Interestingly, they form clusters of about 1µm in diameter, which can look like grapes when stained (2). However, unstained, S. aureus present as golden colonies (2).
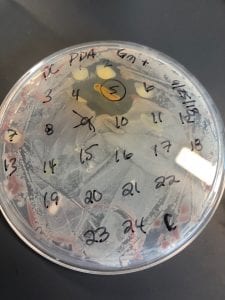
The optimal growing temperature for S. aureusis 37C, while it will grow between 7 and 48C and can survive in temperatures of below -20C (4). Unlike many other bacterium, S. aureus is resistant to high salt content(4). Additionally, S. aureus has the ability to grow under aerobic and anaerobic conditions, although aerobic conditions are preferred(4). Typical medias for colony growth include, blood agar. Tryptic soy agar, and heart infusion agar (5).
As mentioned earlier, S. aureus is typically found in the mucous membranes and on the skin of humans, and are found particularly in the nasal passage (5). However, S. aureuscan also be found in food, and due to its ability to survive in harsh conditions S. aureus is a source of food poisoning (4).
- Clinical Importance and Prevalence
S. aureus infections have stabilized in numbers since the 1990s, with about 10-30 cases per 100,000 people per year (1). However, the number of MRSA diagnosis has been rapidly increasing over the same time period (1). Both community and health care related epidemics have contributed to the increasing number of MRSA outbreaks (1). Newer more intense infection control procedures have helped to reduce the number of outbreaks in recent years (1).
- Infection (How does the infection occur and where is it localized?)
Infection is most common in people at the “extremes of life,” i.e. infants and the older populations (1). Additionally, humans working in the health professions, hospitalized people, and individuals that are immunocompromised are at a greater risk of contracting the infection (2). S. aureusis transmitted via direct contact or fomites (objects that have the bacteria on them such as clothing, tools, furniture) from person to person (2). Infections can occur in virtually every part of the body, however are most common in skin and soft tissues.
- Pathology (What disease is caused? What are the symptoms?)
Many different diseases can be the result of S. aureus. These include, endocarditis, osteomyelitis, septic arthritis, gastroenteritis, meningitis, toxic shock syndrome, urinary tract infections, pneumonia, but the most common being skin and soft tissue infections (2). Depending on the strain and location of the infection as well as the resulting disease, symptoms vary greatly. However, for ease of answering the question, the symptoms for some of the diseases are as follows:
Skin infections (abscess and cellulitis) / MRSA : Swollen, painful bumps that are warm and full of pus, and associated with a fever. Untreated, these lead to deeper more painful infections and can burrow into the skin (6).
Osteomyelitis: fever/chills, swelling of the infected limb with redness, and eventually stiffness and inability to maneuver infected limb. Diagnosis involves blood tests, and potentially a bone scan (7).
Pneumonia: Difficulty breathing, fever, chills, cough. This is diagnosed via chest x-ray and usually involves treatment with antibiotics and hospitalization (8). (Ironic because S. aureus pneumonia is usually transmitted in hospital settings)
- Ineffective Antibiotics (Antibiotics to which the organism has acquired resistance)
Depending on the strain, S. aureus is resistant to a variety of antibiotics (9). The emergence of antibiotic strains began in the 1950s with penicillin resistance (9). Then, in the 1960s, the first strains of methicillin resistant S. aureusemerged (9). And more recently, vancomycin resistant strains emerged (9). Thus, S. aureuscan be resistant to the majority of antibiotics, and discovery of new treatments is ever so important.
- Effective Antibiotics (Antibiotics known to inhibit the organism)
Treatment of S. aureusis usually done with penicillinase-resistant beta-lactams (5). Penicillin is the typical antibiotic of choice for sensitive strains (2). However, due to the many different strains ofS. aureus, characterization of the bacteria may be necessary before treatment. For example, the MRSA strains are resistant to beta lactams and are typically treated with vancomycin. Additionally, other treatments may coincide with the antibiotics such as fluid replacement.
- Corresponding Safe Relative
Staphylococcus epidermidis
Reference:
- Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., & Fowler, V. G. (2015). Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management.Clinical Microbiology Reviews, 28(3), 603–661. http://doi.org/10.1128/CMR.00134-14
- Taylor TA, Unakal CG. Staphylococcus Aureus. [Updated 2017 Oct 9]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2018 Jan-.Available from: https://www.ncbi.nlm.nih.gov/books/NBK441868/
- Green, B. N., Johnson, C. D., Egan, J. T., Rosenthal, M., Griffith, E. A., & Evans, M. W. (2012). Methicillin-resistantStaphylococcus aureus: an overview for manual therapists. Journal of Chiropractic Medicine, 11(1), 64–76. http://doi.org/10.1016/j.jcm.2011.12.001
- https://www.foodstandards.gov.au/publications/Documents/Staphylococcus%20aureus.pdf
- https://www.ncbi.nlm.nih.gov/books/NBK8448/
- Chambers, H. F., & DeLeo, F. R. (2009). Waves of Resistance:Staphylococcus aureus in the Antibiotic Era. Nature Reviews. Microbiology, 7(9), 629–641. http://doi.org/10.1038/nrmicro2200
- https://www.mayoclinic.org/diseases-conditions/mrsa/symptoms-causes/syc-20375336
- https://www.healthline.com/health/osteomyelitis
- http://www.health.state.mn.us/divs/idepc/diseases/staph/basics.html