PCR resulted in successful amplification of two of my three isolates of interest (#5 and #11). Although #18 inhibited both Gram positive and Gram negative tester strains, BLAST sequence analysis could not be done to determine the genus, because no DNA was amplified to sequence! Additionally, neither Gram staining nor extraction were performed for isolate #5. Thus, here I identify and discuss the genus of isolate number 11 for which Gram stains and BLAST were both successfully performed.
What were the results of your 16S analysis?
BLAST retrieved a list of organisms (genus and species) that contain a high level of sequence similarity in the DNA region coding for the 16S rRNA subunit (693 base pairs analyzed). The results are seen below… and my microbe is…Bacillus! The genus Bacillus dominated the sequence similarity results in both BLAST and the Ribosomal Database Project (RDP) (results seen below). Both proposed a list of species and strains within the Bacillus genus that demonstrated similarity in 16S sequence. Bacillus subtilis was the most common species to appear in these lists, in a variety of strains. This is interesting because Bacillus subtilis was one of our safe-analog ESKAPE tester strains! A few such proposed strains were Bacillus subtilis NBRC 13719, Bacillus subtilis DMS 10, and Bacillus subtilis IAM 12118. However, Bacillus strain identification is limited to the species level in 16S rRNA analysis (1).
Another species that was listed as similar was Bacillus licheniformis, which has strains known to produce antibiotics such as bacitracin and bacteriocin (1). Since isolate 11 inhibited tester strains, it is possible that it was Bacillus licheniformis. Nevertheless, 16S results definitely point to Bacillus.
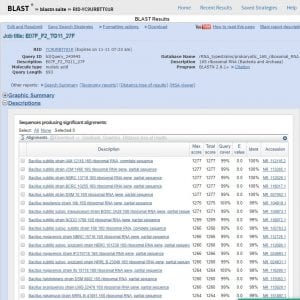
BLAST results listing organisms with highly similar sequences of DNA coding for 16S rRNA. Genus and species of similar organisms are included.

RDP results, affirming the genus as Bacillus in accordance with BLAST. Bacillus subtilis appears often in the list of similar strains, as it did in BLAST. This suggests isolate 11 may be a strain of Bacillus subtilis.
Does your gram stain agree?
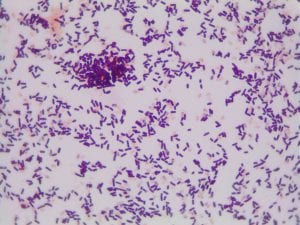
YES! My Gram stain showed that isolate 11 was Gram positive rods and upon close examination it appeared that they formed spores. We also know that this isolate must have been aerobic since it grew in our aerobic conditions.
The genus of Bacillus contains Gram-positive, spore-forming, rod-shaped, aerobic bacteria (1). All of this aligns with what we know to be true about my isolate 11! Thus, I feel comfortable claiming that isolate 11 is some species of Bacillus.
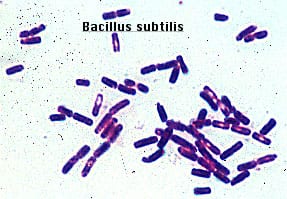
Gram positive rods, just like isolate 11. Form endospores which resemble the spore-looking characteristic of isolate 11. https://www.medschool.lsuhsc.edu/Microbiology/DMIP/dmex17.htm
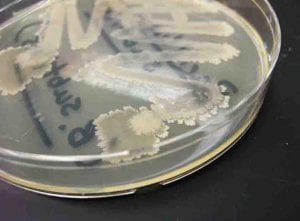
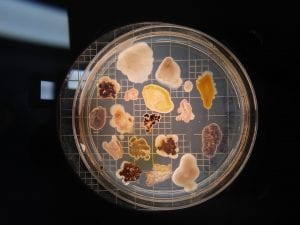
This streak plate of Bacillus subtilis found online resembles closely the morphology of my isolate 11 streaks in particular the color and unique pattern of growth on the edges of colonies. https://www.scienceprofonline.com/science-image-libr/sci-image-libr-bacterial-colonies.html
a) General cellular and morphological characteristics of the genus (taxonomic classification, nutrition, cell shape, habitat).
Taxonomic classification. Domain: Bacteria; Phylum: Firmicutes; Class: Bacilli; Order: Bacillales; Family: Bacillaceae 1; Genus: Bacillus
These are Gram-positive, spore-forming, rod-shaped, aerobic bacteria (1). They are widely distributed in the soil as well as the aquatic environment (2). Their ability to inhabit diverse habitats results in part due to their spore forming ability (2). Some Bacillus species inhabit extreme environments such as high temperature or extreme pH since spores can survive dormant in extreme environmental conditions (3). Bacillus can also survive in low-nutrient environments, including a lack of elements such as phosphorus, nitrogen or oxygen, provided that the organism has a sufficient supply of carbon sources (1). Bacillus represent heterogeneous species that also form bioactive polymers useful in industry including medicine, biodefense, biofuels, and bio-pesticides. For example, Bacillus thuringiensis is used for biological control of insects to protect crops (1). Spores of Bacillus subtilis have also been used as probiotics in humans (1).
b) Information regarding antibiotic production in this genus.
The number of antibiotics produced by the genus Bacillus is in the hundreds (4). Many of these antibiotics are peptide antibiotics, which tend to be smaller than proteins but range in molecular weights from 270 (bacilysin) to 4500 (licheniformin) (4). 66 different peptide antibiotics are elaborated by strains of Bacillus subtilis (4) and 23 by Bacillus brevis (4). Most of these peptide antibiotics are made exclusively from amino acids, but some contain additional substituents (4). As mentioned above, the species Bacillus licheniformis is known to contain strains that produce antibiotics including bacitracin and bacteriocin (1). Additionally, polymyxin and gramicidin S have been isolated from species of Bacillus. Antibiotics of Bacillus have been seen to be more effective against Gram positive bacteria (4), which is consistent with my observations of isolate 11.
References
(1) Porwal S., Lal S., Cheema S., Kalia V.C. (2009). Phylogeny in Aid of the Present and Novel Microbial Lineages: Diversity in Bacillus. Plos One.
(2) Parvathi A., Krishna K., Jose J., Joseph N., Nair S. (2009). Biochemical and molecular characterization of Bacillus pumilus from coastal environment in Cochin, India. Brazilian Journal of Microbiology. 40(2):269-275.
(3) Nicholson W.L., Munakata N., Horneck G., Melosh H.J., Setlow P. (2000). Resistance of Bacillus endospores to extreme terrestreial and extraterrestrial environments. Microbiology and Molecular Biology Reviews. 64(3):548-572.
(4) Katz E. and Demain A.L. (1977). The peptide antibiotics of Bacillus: Chemistry, Biogenesis, and Possible Functions. Bacteriological Reviews. 41(2):449-474.