What were the results of your 16S analysis?
The 16S analysis results showed the genus of my species is Bacillus. The closest relative of my bacteria, determined by BLAST, is Bacillus subtilis. This relative had a percent identity of 99% and the most common sequence alignment. The phylogenetic tree produced by BLAST explains that other relatives with high percent identity values are close together in the tree. For example, Bacillus glycinifermentans, Bacillus haynesii, and Bacillus sonorensis are all of 99% identity values, but had less similar “hits” or or sequence comparisons than B. subtilis.
Figure 1. Phylogenetic tree, produced by BLAST, showing the relationship between bacteria that produce 99% identity matches. Although they produce similar % identities to B. subtilis, they have less overall sequence similarities to my unknown sample.
Does your gram stain agree?
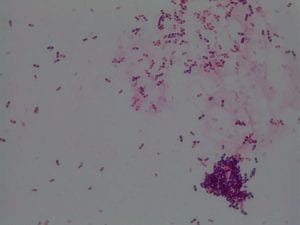
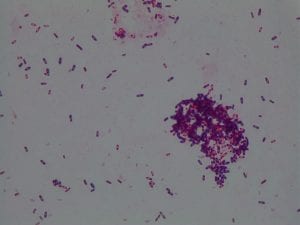
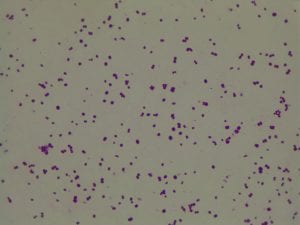
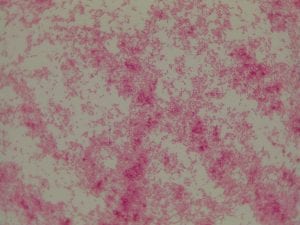
My gram stain agrees! Bacillus is Gram positive, rod-shaped, spore forming bacterium. My Gram stain shows all of these features. The image below shows the Gram stain of my bacteria. The rods are purple in color with a pink center. The purple color signifies a Gram positive bacteria. The pink center shows that the bacteria is spore-forming.
Pick one of your isolates and find out more about the genus (it is unlikely you will be able to determine the species).
I only have one isolate, so that is the one that I have picked (KM5). The genus is Bacillus.
a) General cellular and morphological characteristics of the genus (taxonomic classification, nutrition, cell shape, habitat).
There are over 200 known species of Bacillus, but only two are known to impact humans. Bacillus can be classified as follows: Kingdom- Bacteria, Phylum- Firmicutes, Class- Bacilli, Order- Bacillales, Family- Bacillaceae. Bacillus are rod-shaped, Gram positive, endospore-forming bacteria. The bacteria that has been isolated in lab, as well as most of the Bacillus genus is aerobic. There are some species that act as facultative anaerobes. Due to most species being aerobic, their nutritional well-being depends on having oxygen. To achieve optimal growth in a lab setting, Bacillus prefers a rich media. With that being said, the genus would grow best in its natural environmental conditions and it is hard to replicate those. Their optimal temperature range for growth and life is between 25-35 degrees Celsius. Finally, the genus are known to inhabit soil. However, they can also be found in aquatic environments. Some species, such as B. subtilis can be found naturally in the human gut.
b) Information regarding antibiotic production in this genus.
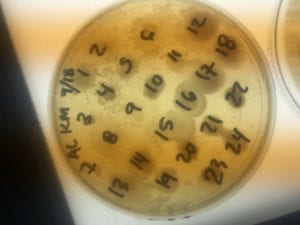
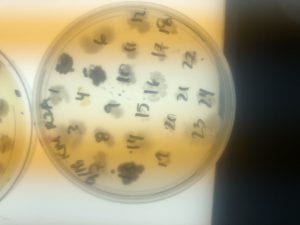
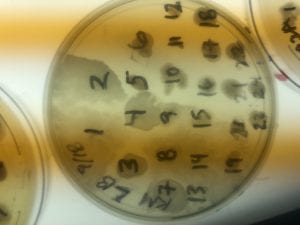
The Bacillus species produces antibiotics. Antibiotics act as the bacteria’s “fight” mechanism. Some examples of common antibiotics that the Bacillus genus excretes are polymyxin, bacitracin, and gramicidin. The antibiotics that the Bacillus genus produces are effective against both Gram positive and Gram negative bacteria. From my experiments, I determined that my isolate was only effective against Gram positive bacteria. This is not unusual for a variety of species of Bacillus. The genus can also have other antimicrobial uses, such as being an anti-fungal.
Sources:
https://www.sciencedirect.com/science/article/pii/S0944501305000704
https://www.ncbi.nlm.nih.gov/books/NBK7699/
https://www.flandershealth.us/microbiology/i-characteristics-of-bacillus-subtilis.html
http://www.heathermaughan.ca/resources/Maughan%26vanderAuwera11.pdf
https://microbewiki.kenyon.edu/index.php/Bacillus_subtilis
https://www.ncbi.nlm.nih.gov/blast/treeview/treeView.cgi?request=page&blastRID=Y38Z9EVS014&queryID=lcl|Query_28503&entrezLim=&ex=&exl=&exh=&ns=100&screenWidth=1280&screenHeight=800