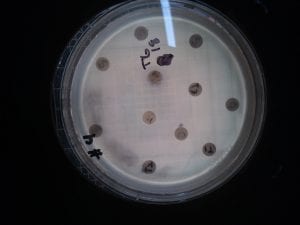
I tried to extract organic molecules from the plate containing the colonies I called v1 3 and v1 9. I tested it against the Gm+ Staph epi and B. subtilis, and the Gm- E. caratavora and P. putida, since one or both of my isolates inhibited each one. There was only a faint zone of inhibition in the B. subtilis from the v1 3 extract. Since the isolates were only grown for one day, it could be that there wasn’t much of any production of secondary metabolites yet.
Category Archives: 6. Extract news!
Extract news! -Carly B
I had two samples, 2 and 4, to perform the extract with. Sample 2 did not evaporate so I could not add methanol to it but sample 4 did evaporate. I performed the agar disc diffusion test with my two extracts against tester strains #3, #4, #6, and #7. Strain #4 was E. raffinosis and #6 was B. subtilis and they were Gm+. Tester strain #3 was E. carotovora and #7 was E. aerogenes and they were Gm-. I chose E. raffinosis and B. subtilis because my samples previously showed inhibition against these two strains and then I randomly chose two Gm- tester strains because my samples did not show previous inhibition of any Gm- tester strains. Extract 2 did not inhibit any of the tester strains. Extract 4 inhibited E. raffinosis and B. subtilis, both Gm+ bacteria, so this sample seems to be an antibiotic producer against Gm+ bacteria.
Extract News! Sofia
From my 3 (4, 6, and 11) different isolates, I obtained 5 different extracts. Isolates 6 and 11 produced two different extracts because the initial extract produced three different layers, two of which were of interest for extraction. To test my extracts for antibiotic production, I chose 2 different Gram positive and 2 different Gram negative strains to use. The Gram positive strains I chose were: S. epidermis (1) and E. raffinosis (4) and the Gram negative strains that I chose were: E. coli (2) and A. baylyi (5). I chose strains 2 and 4 because when we previously tested our isolates against the ESKAPE pathogens, I had observed inhibition of all three of my isolates against these two strains. I chose 1 and 5 because there had been slight signs of inhibition when previously tested, which may have become more prominent with a prolonged growth time.
Unfortunately when my extracts were tested against each of these strains, none of my extracts displayed antibiotic-producing activity against any of the tester strains. To further conclude that these bacterial isolates are not antibiotic producers, I could test the extracts against different strains, or test them on a different type of media. Additionally, the plates could be left to grow for a longer period of time. If the observations continued to be the same, it would be reasonable to conclude that none of my isolates are antibiotic producers.
Extract News! Troy
Of all the fabulous bacteria that I had, there were three strains that were worthy of testing. Coincidentally, all three were grown on R2A – a reason for which may be that there were fewer resources in this substrate. The extracts were gathered on Thursday, October 25, by collecting the organic layer of our previously suspended colony plates. I extracted three different ‘antibiotics’ from my three different strains, and prepared them to be dried. On October 30, the extracts (which were not completely dried yet) were prepared to be tested against different bacteria strains. They were suspended in methanol, and this solution was used to create antibiotic plates that were dropped onto the different tester bacteria plates. I chose to test against 4 strains – 3, 4, 6, and 7 -, as these strains presented two GM(+) and two GM(-) bacteria strains. On November 1, the results were observed, and of my three extracts I had antibiotic activity against both GM(+) and GM(-) bacteria. Strain 1A – a GM(-) bacillus – produced antibiotics that inhibited growth on bacteria strain 3 – a GM(-) bacteria. Strain 2B – another GM(-) bacillus – produced antibiotics that inhibited bacteria strain 7 – a GM(-) bacteria. Strain 3A – a GM(-) rod-shaped bacteria – produced antibiotics that inhibited bacteria strain 4 – a GM(+) bacteria.
I was very happy to see that I had AB production from all three of my strains. I was interested that the two bacilli strains produced AB’s that inhibited the GM(-) testers while the rod-shaped strains produced AB’s that inhibited the GM(+) tester strain (strain 4). I believe that these strains could be further observed to be potential drugs against bacteria, as they target a wide range of GM(+) and GM(-) bacteria.
Extract News! Kara
I tested 3 extracts from my 3 producers, one of which was isolated on PDA, while the other 2 were isolated on 10% TSA. I found that all three of my extracts inhibited strain #1 which was staph epi, but did not inhibit the other strains. I picked the strains a bit arbitrarily, because one of my producers had inhibited all of the strains, so I chose 2 gram negative, and 2 gram positive. I chose strains 1 (Staph epi), 2 (E. coli) , 5 (A. baylyi) and 6 (B. subtillis). I made sure A. baylyi was one of the strains I looked at because my ESKAPE pathogen was A. baumanni (which is the pathogenic relative of A. baylyi), so I was curious as to whether or not my extracts would inhibit it (sadly they did not).
Overall, I was happy to see that my extracts demonstrated some antibiotic activity, indicating that my extraction was successful. Unfortunately, staph epi is very “easy” to inhibit, indicating my extracts probably would not work against most microbes.
Extract news! – Tommy
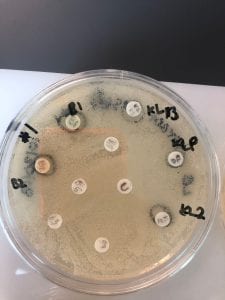
On October 25th, we performed an organic extraction of our isolates. I had two isolates that I attempted extractions on. After letting the extraction vials dry, a residue was present which was weighed and subsequently resuspended in methanol. Not all of the residue dissolved, but 20 microliters of this resuspended methanol/extract solution was put on little circles to put on tester strains to see if they inhibited tester strains. If the tester strains were inhibited, it would mean we were successful in extracting an antibiotic from the bacteria!
We tested our extractions on two Gram positive and two Gram negative tester strains. Both of my isolates had inhibited at least one Gram positive and one Gram negative tester strain. To test extractions, I chose testers 1, 3, 4, and 5 (Staphylococcus epidermidis, Erwinia carotovura, Enterococcus raffinosus, and Actinetobacter baylyi, respectively). Testers 1 (Gram positive) and 5 (Gram negative) were previously inhibited by isolate number 11 and testers 3 (Gram negative) and 4 (Gram positive) were previously inhibited by isolate number 18.
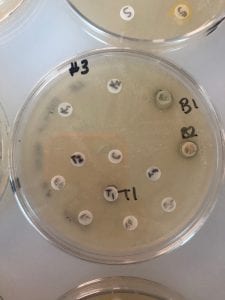
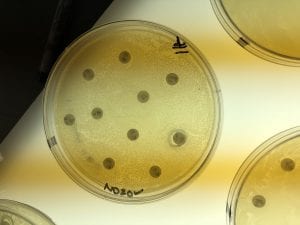
Below are pictures of the results of testing the extracts on testers 3 and 4 (Erwinia carotovura and Enterococcus raffinosus). Extract from isolate number 18 inhibited both testers as seen by the clearing of growth of the tester bacteria around the 18 extract circle. This is interesting because I chose these two testers based on inhibition that I saw from isolate number 11, not isolate 18, but extract 18 showed inhibition while isolate 11 did not. However, extract 18 also inhibited testers 1 and 5 (Staphylococcus epidermidis and Actinetobacter baylyi), albeit not as robustly. Extract 11 did not show inhibition of any tester strains. The fact that extract 18 inhibited tester strains indicates that an antibiotic was successfully extracted!
Tester 3 – Erwinia carotovura – with organic extractions. The extraction from my isolate 18 shows a clearing of growth around it, indicating inhibition of the tester.
Extract News- Kelsey
I tested two extracts. One extract was an isolate of mine on 10% TSA and the other extract was the same isolate on R2A. I tested the extracts on two Gram negative strains, E. carotova and E. coli. I also tested the extracts on two Gram positive strains, E. raffinosis and B. subtilis. When testing the extracts, I found no resistance observed to any of the strains. When the two isolates were tested, they had observed resistance against E. raffinosis and B. subtilis. If I were to further test the extracts, I could try different media or the same media again. I could also leave the plates out for longer to see if any antibiotic activity would show over time. However, at this point, I conclude that the isolates did not secrete any antibiotic activity into the media on which they were plated.
Extract News! – Le Minh
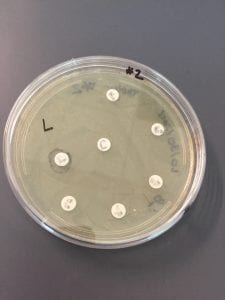
I was able to obtain an extract from the only isolate (LB #5) that I had. The extract appeared insoluble in methanol as I had a hard time resuspending it; the extract formed a large white clump that would not fully dissolve even after lengthy vortexing. However, I still carried out the following procedure of testing the extract with the obtained solution. I chose to test it against the Gram-negative #2 E. coli and #7 E. aerogenes as well as against the Gram-positive #4 E. raffinosus and #6 B. subtilis as my isolate was able to grow in those tester strains during the ESKAPE testing we performed before.
In the end, my extract did display some antibiotic-producing ability (so the extract was somewhat successfully resuspended). Out of all test plates, my extract was able to show inhibition only against E. coli but not against the other Gram-negative bacterium nor the Gram-positive bacteria. This means that my extract has narrow-spectrum antibiotic activity.
The picture below shows the zone of inhibition around my extract (it is a little bit hard to see but it is there) that is was able to produce against E. coli
Extract News! -Alexa
I tested four of my bacteria samples (13, 14, 15, and 17) against the tester strains 1 (S.Epidermis),3 (E. Carotovora) ,6 (B. Subtillis), and 9 (P. Putida). I chose these strains because my bacteria had previously shown resistance to all of them. Out of my four bacteria, only one of them, number 15, showed inhibition against strain 6. It had a very small ring around it so it didn’t show strong inhibition. Previously, number 15 did show inhibition against tester strain number 6. Number 13 and 14 both came from my LB plates and showed inhibition against tester strains 1, 3, and 6, so it was surprising that I didn’t see an inhibition from the extracts against any of the tester strains. Number 17 was from my PDA plates and showed inhibition against strains 6 and 9, but its extract also failed to show inhibition against any of the strains. Since only one of my samples showed inhibition it could be possible that I didn’t get extracts for the other ones or maybe the concentration was so low that it didn’t make a difference. It could have also been helpful to incubate the tester strains with the extracts on them longer, or let the heavy streak plates incubate longer since it’s possible the bacteria needed a few more days to start producing an antibiotic.
Extract News! – Brianna
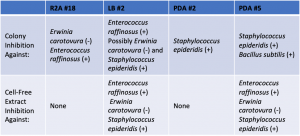
I was able to obtain a total of 6 extracts between my 4 samples (there was a little bit of labeling issues, so I could not combine the duplicate sets). I chose to test these extracts against #1 (Staphylococcus epideridis) and #4 (Enterococcus raffinosus) as the gram positive ESKAPE pathogens and #3 (Erwinia carotovura) and #7 (Enterobacter aerogenes) as the gram negative. These were chosen since my producers showed inhibition against these pathogens (some showed against one or two, but not all three at once) with the exception of number 7 (which was chosen out of convenience).
Out of all the extracts, two showed inhibition against Staphylococcus epideridis, Enterococcus raffinosus and Erwinia carotovura . While the numbering/labeling was messed up from before, these were my two pigmented extracts. I believe these correspond to my samples, PDA #2 and LB #2.
Here is a table summarizing the cell free and colony inhibition tests.
Extract News! – Elizabeth
The last day of my extract procedure was completed by Professor Salvo, and thus she picked the tester strains that were used. Upon inspection of the plates, no extracts were successfully isolated; none of the disks had zones of inhibition around them. This could mean that the ethyl acetate procedure wasn’t effective for these compounds, or something else could have gone wrong with the procedure. If we had more time, I would redo the extraction with methanol instead to see if the ethyl acetate was the problem.
Example plate showing no zones of inhibition.
6. Extract News! – Nikki
Did you obtain an extract? Summarize the results of your testing and why you choose the tester strain(s) you did.
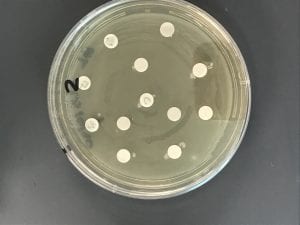
I was able to obtain an extract for each of my three isolates (#15, 20, and 23) following the extraction procedures we followed over the last few weeks in lab. I chose to use the following tester strains: #1 S. epidermidis (+), #2 E. coli (-), #6 B. subtilis (+), and #7 E. aerogenes (-). I chose these four tester strains because my isolates had shown inhibition previously of #1, #2, and #6 tester strains and then I chose #7 because it was a gram – strain to fulfill the 2 Gram + and 2 Gram – that we testing against in this stage. In my observations from the test extracts, I only saw a ring signaling potential inhibition around my #20 isolate/extract on the #1 S. epidermidis, a Gram + tester strain. There was no other antibiotic activity on the other plates/for the other extracts. I added a photo below of the #20 isolate on the #1 tester strain plate and it is the one circled and labeled “ND 20”. Note that this photo was taken on Thursday, November 1st, a day after Prof. Salvo took the plates out of the incubator and made the observations of antibiotic activity.