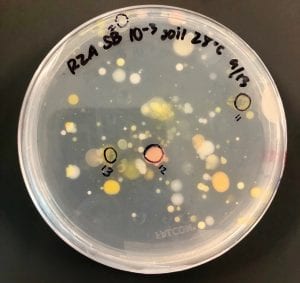
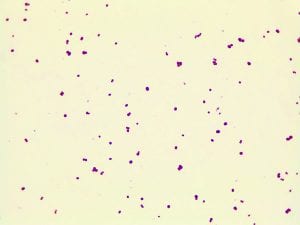
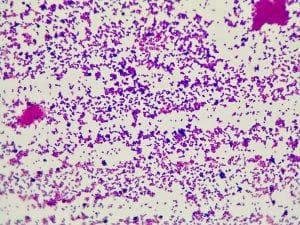
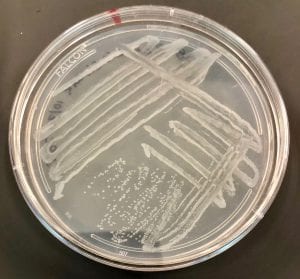
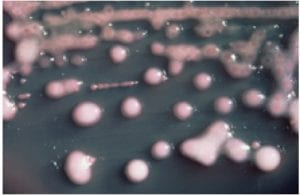
Two of my original plates with my serial dilutions from my soil sample. I plates my dilutions on R2A, LB, AC, and 10% TSA. I had the widest variety of shape, size and color on R2A. With later tests I also have the best growth, and the most inhibition on R2A, so the majority of later tests were done using R2A plates.
My second patch plate of the 15 colonies picked from my original R2A plates. Patches 4, 6, and 11 exhibited signs of inhibition. These are the isolates I used for further testing.
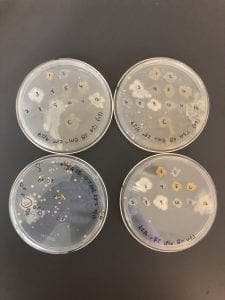
LB plates: top two are the plates used for the first test against the ESKAPE pathogens; no inhibition. Bottom left is my serial dilution plate. Bottom right is Patch plate #2.
10% TSA plates: top two are the plates used for the first test against the ESKAPE pathogens; no inhibition. Bottom left is my serial dilution plate. Bottom right is Patch plate #2. Similar growth to the R2A plates, however nothing that was patches exhibited any signs of inhibition.
AC plates: top two are the plates used for the first test against the ESKAPE pathogens; no inhibition. Bottom left is my serial dilution plate. Bottom right is Patch plate #2.
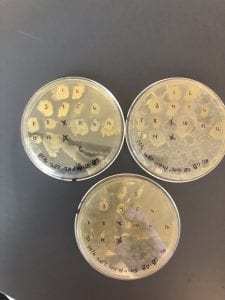
Second set of LB plates used to test against a Gm +/- strain of a “safe” ESKAPE pathogen. Lots of slimy/ gooey yellow growth, but no signs of inhibition.
Second set of AC plates used to test against a Gm +/- strain of a “safe” ESKAPE pathogen. Also had lots of slimy/ gooey yellow and orange growth and the patch plates had very large and funky shaped patches; no signs of inhibition.
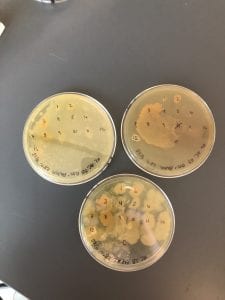
R2A plates with patches to test against Gm +/- strains of “safe”ESKAPE pathogens. There was not much inhibition against the Gm+ strain, but there was signs of inhibition against the Gm – strains. This is consistent with the results of the second ESAKPE pathogen test, in which we tested against all 8 strains. Signs of inhibition were more commonly seen against the Gm – strains.
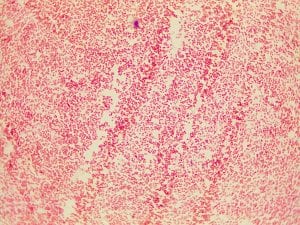
Gram stain for the Gm + control (S. epidermis)
Gram stain for the Gm – control (E. coli)
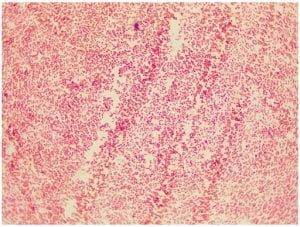
Gram stain for isolate #4; Gm -; appears to be a small cocci but further examination concludes that the bacteria is a very, very small and thick rod, that appears somewhat round.
Gram stain for isolate #4; Gm (purple in color); small cocci, some are diplococci and there are a few short streptococci (chains).
Gram stain for isolate #11; Gm – (pink in color); appears to be a small cocci but further examination concludes that the bacteria is a very, very small and thick rod, that appears somewhat round.
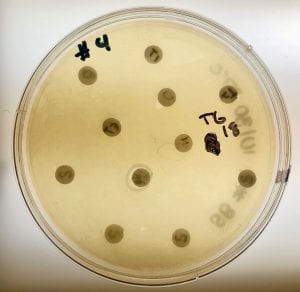
One of my plates from the extract test; My three isolates produced five different extracts, however none of them exhibited signs of inhibition. This is indicates that none of my isolates are antibiotic producers. Signs of inhibition may possibly be visible if incubation time was longer. Additionally, it is possible that something went wrong with the extract, which is why there was no visible inhibition. A repeat of this test could provide more definitive conclusions.
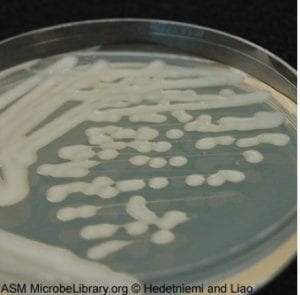
Streak plate for isolate #6 with visible single, isolated colonies.