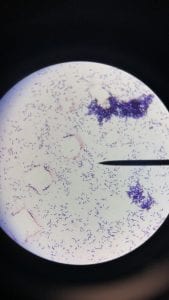
Assigned ESKAPE Pathogen: Klebsiella pneumoniae
Why is this ESKAPE Pathogen of interest (in brief)
Klebsiella pneumoniae is a common nosocomial (originating in a hospital) pathogen that accounts for about 10% of all infections acquired within a hospital setting (1). It is also a common pathogen responsible for community-acquired infections, as well as a number of other diseases such as meningitis, septicemia, purulent abscesses, and pneumonia (1). Additionally, as some strains are nitrogen-fixing, K. pneumoniae is of agricultural interest because it has been observed to increase crop yields in certain agricultural conditions.
General Cellular and Morphological Characteristics of the Organism (taxonomic classification, nutrition, cell shape, habitat)
K. pneumoniae is a Gram negative, non-motile, lactose-fermenting, facultative anaerobic, rod-shaped bacterium. It is a part of the Enterobacteriacae family (6). K. pneumoniae is commonly found in the normal flora of the mouth, skin, and intestines, but can become destructive and cause damage if inhaled, specifically to the alveoli. K. pneumoniae occurs naturally in the soil and about one third of strains are nitrogen-fixing in anaerobic conditions.

https://chrislima90.weebly.com/blog/top-advice-on-klebsiella-pneumoniae
 http://faculty.ccbcmd.edu/courses/bio141/labmanua/lab12/Mac_kpneumoniae.html
http://faculty.ccbcmd.edu/courses/bio141/labmanua/lab12/Mac_kpneumoniae.html
(Grows very well on MacConkey agar)
Clinical Importance and Prevalence
K. pneumoniae originated in a hospital setting and is now globally prevalent in hospital and community settings (6). Infections are more common in the young, the old, and the immunocompromised (6). Enterobacteriacae are becoming more and more difficult to treat because of their increasing resistance to antibiotics through the production of different enzymes such as extended-spectrum beta-lactamases and carabapenemases (6).
Infection (How does the infection occur and where is it localized?)
K pneumoniae infections are typically seen in individuals with a compromised immune system, and most infections are seen in middle-aged or older men who already have debilitating diseases. Healthy people typically do not get Klebisella pneumonia infections (3). Humans serve as the primary reservoir for this pathogen, and the pathogen is typical transmitted between individuals through fecal matter and nasal secretions. K. pneumoniae is rarely carried on the skin (2). Carrier rates of the pathogen increase significantly in hospital patients (2). Klebisella pneumonia is spread through contact, either person-to-person contact (i.e. touching the hands of an infected individual) or environmental contact (i.e. touching a contaminated surface). The bacteria cannot be spread through the air (3). Certain medical tools such as ventilators and catheters may cause an individual to be exposed as well (3). K pneumoniae are capable of infection because of fimbrial adhesins and a thick capsule that is comprised of two layers of polysaccharide fibers (6).
Pathology (What disease is caused? What are the symptoms?)
The most common disease caused by Klebisella pneumoniae is pneumonia. This occurs when Klebisella bacteria enter the respiratory tract and setting in the air sacs of the lungs, causing infection and inflammation (3). The most common symptoms of pneumonia are fever, cough, chest pain, difficulty breathing, and abnormal mucus production (4). Without treatment, pneumonia can become a very serious infection, especially in the elderly and those who are immunocompromised (4). Other diseases that can be caused by Klebisella pneumonia are septicemia, meningitis, endocarditis, and cellulitis (4).
Ineffective Antibiotics (Antibiotics to which the organism has acquired resistance)
Some K. pneumonia have become very resistant to a class of antibiotics called carbapenems (3). Resistant bacteria produce an enzyme called carbapenemase which renders carbapenems ineffective (3). These resistant bacteria are also known as KPC-producing organisms and are very difficult to treat (3).
Effective Antibiotics (Antibiotics known to inhibit the organism)
There are many effective antibiotics to treat K. pneumoniae (5). Beta-lactams, aminoglycosides, and quinolones are effective antibiotics against Klebsiella infections (5). Cephalosporins have also been used alone and in conjunction with aminoglycosides but should not be used if ESBL strains are present (5).
Corresponding Safe Relative
The corresponding safe relative is Escherichia coli (6). Most E. coli are commensal and make up about 0.1% of the of the normal intestinal flora, however some are pathogenic and has shown marked resistance to multiple antibiotics in the past decade (6).
Sources:
1. Guo, S., J. Xu, Y. Wei, Y. Li, and R. Xue. 2016. Clinical and molecular characteristics of Klebsiella pneumoniae venitllaor-associated pneumonia in mainland China. BMC Infec Dis. 16: 608
2. Yu, W.L., Y.C. Chuang. UpToDate. Clinical features, diagnosis, and treatment of Klebsiella pneumoniae infection. 2018. https://www.uptodate.com/contents/clinical-features-diagnosis-and-treatment-of-klebsiella-pneumoniae-infection
3. Centers for Disease Control and Prevention. Klebsiella pneumoniae in Healthcare Settings. 2012. https://www.cdc.gov/hai/organisms/klebsiella/klebsiella.html
4. WebMD. What is Klebsiella Pneumoniae Infection?. 2018. https://www.webmd.com/a-to-z-guides/klebsiella-pneumoniae-infection#1
5. S. Qureshi. Medscape. Klebsiella Infections Medication. 2017. https://emedicine.medscape.com/article/219907-medication#showall
6. Pendleton, J. N., Gorman, S. P., & B. F. Gilmore. Medscape. Clinical Relevance of the ESKAPE Pathogens. 2013.