What were the results of your 16S analysis?
| Isolate #/ Name
(Include # bp) |
Closest Relative | Identity
(%) |
Query Coverage (%) | Accession Numbers |
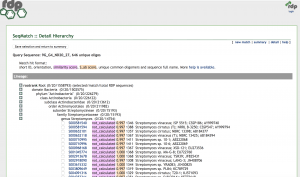
| #20
(677 bp) |
Streptomyces
(vinaceus or cirratus strains) |
99 | 99 | NR_041131.1
NR_112388.1 |
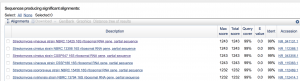
Unfortunately, I was only able to obtain 16S results for one of my isolates, #20, as the other two, #15 and #23 did not work. However, for #20, I was able to run 677 bp of reliable sequence in my Blast and Ribosome Database Project (RDP) searches. Both searches agreed on Streptomyces genus as the identify of my isolate with 99% identity match and 99% query coverage. Further, my searches determined the closest relative to be Streptomyces of the vinaceus or cirratus strains, but more informations and biochemical tests would be needed to distinguish any further.
Does your gram stain agree?
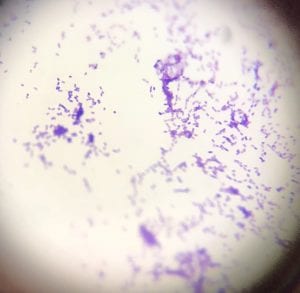
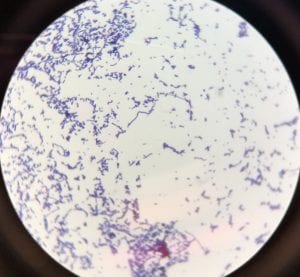
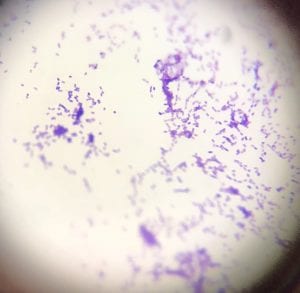
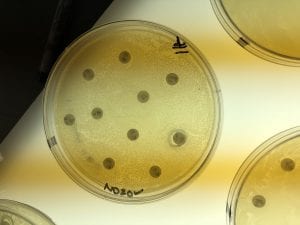
This is a photo of my gram stain for isolate #20 (photos of controls and other info in my “Meet My Microbes!” post). From my gram stain, I determined my isolate to be Gram + with a rod-shape/bacillus and filamentous microscopic morphology. Therefore, my gram stain agrees with my 16S data because Streptomyces is a genus of Gram positive bacteria with a filamentous shape [1].
Pick one of your isolates and find out more about the genus (it is unlikely you will be able to determine the species).
a) General cellular and morphological characteristics of the genus (taxonomic classification, nutrition, cell shape, habitat).
b) Information regarding antibiotic production in this genus.
In terms of taxonomy, Streptomyces is the genus of the Streptomycetaceae family, which is further broken down into over 575 species, with more and more still being discovered. Additionally, Streptomyces belongs to the Bacteria Kingdom, Actinobacteria Phylum and Class, and Actinomycetales Order [2]. Streptomyces is known to be Gram positive bacteria that can grow in various environments, and has a filamentous morphology [1]. In their simplest form, Streptomyces are unicellular spheres and rods and are filamentous. However, in more complex forms, they are described as “mycelium of branching hyphal filaments, and reproduce in a mould-like manner by sending up aerial branches that turn into chains of spores” [3]. Streptomyces are primarily found in soil and decaying vegetation, which further confirms that this could be my isolate #20 because I isolated it from my soil sample from a garden [2, 4]. Interestingly, the first Streptomyces are thought to date back 400 millions years ago to when Earth last was first colonized by plants and as such their job was to solubilize the cell walls or other other components of plants, fungi, and insects. This function is further thought to be retained as certain species have genes for proteins that are involved in the binding and degradation of certain carbohydrates[4]. In terms of nutrition, the genus is primarily found in soil and so upon studies for optimum growth of Streptomyces in media, it was determined that various trace elements proved to be important including copper, manganese, and zinc, as well as using sodium nitrate, potassium phosphate, and magnesium sulphate [5].
In today’s society, Streptomyces are of utmost importance as a major source of antibiotics in medical, veterinary, and agricultural use [3]. Further, Streptomyces best known for their ability to produce bioactive secondary metabolites including antifungals, immunosuppressants, and perhaps most importantly in our case, antibiotics. The production of antibiotics is species specific and are important for them to compete with other microorganisms they meet [1]. Finally, the importance of this genus is expressed in the statistic that 80% of antibiotics commonly used today come from the Streptomyces genus [1].
[1] De Lima Procópio, Rudi Emerson, et al. “Antibiotics produced by Streptomyces.” Antibiotics produced by Streptomyces. https://www.sciencedirect.com/science/article/pii/S1413867012001341
[2] Streptomyces, Wikipedia. https://en.wikipedia.org/wiki/Streptomyces
[3] “Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics” Philosophical transactions of the Royal Society of London. Series B, Biological sciences vol. 361,1469 (2006)
[4]. “Recent advances in understanding Streptomyces” F1000Research vol. 5 2795. 30 Nov. 2016, doi:10.12688/f1000research.9534.1
[5] Spilsbury, J. F. “Observations on the nutritional requirements of Streptomyces griseus (Krainsky) Waksman & Schatz.” Transactions of the British Mycological Society.