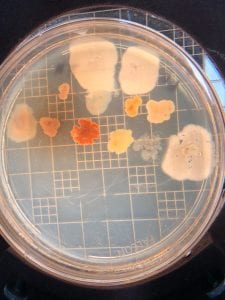
First, bacteria were grown on three different culture media – LB, AC, and R2A – at three different dilution factors. LB and AC were grown at 10-2, 10-3, and 10-4, while R2A was grown at 10-1, 10-2, and 10-3. This was done, because they provided three different nutrient levels; Rich, somewhat rich, and somewhat sparse.
After the bacteria were grown they were selected for different traits that set them apart. They were then picked and patched onto individual media corresponding to the media they were picked from. 12 cultures were picked from LB, 8 were picked from AC, and 16 were picked from R2A.
I thought it was so cool how many different colors and textures were produced by these bacteria! It’s safe to say that Professor Salvo was right – this soil is pretty diverse!
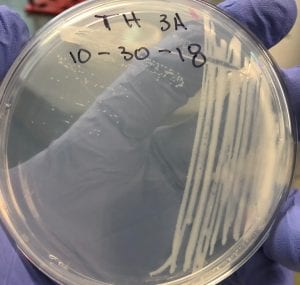
After testing all of these strains against different tester bacteria, the only colonies that actually exhibited any inhibition pattern were strains 1A, 3A, and 2B. These colonies were isolated after they were tested, and even though I started off a little rough with streak plates I think it’s safe to say I got the hang of them by the end!
Just look at those beautiful streaks! Gotta bring these bacteria over to the Nott… (insert laughs here)
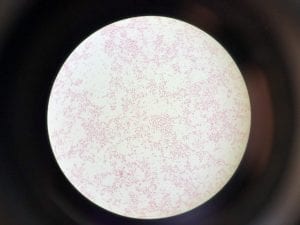
So you might be asking yourself, what kind of bacteria am I actually looking at here? Have no fear! After we isolated these strains we used a Gram staining method to determine if our bacteria were Gram positive of Gram negative. If the bacteria were purple they were Gram positive, and if the bacteria were pink they were Gram negative.
So you might be asking yourself, “Did these bacteria actually produce any antibiotics?” And your answer is YES! They did! After testing all of the antibiotics against four different strains of bacteria it was revealed that they each inhibited at least one strain!
If you look at the individual plates (in the red circles) you’ll see that there are little halos around them! These halos are the proof that the antibiotics worked, as they successfully killed off the bacteria! While these rings are small it is still super cool that the antibiotics actually worked. Hopefully in the future someone will be able to determine the mechanism by which it works, and enhance the function of the antibiotic or further the search for antibiotics in these types of bacteria! Who knew a little soil sample from outside the construction zone would be so interesting…