Assigned ESKAPE Pathogen: Pseudomonas aeruginosa
Why is this ESKAPE Pathogen of interest
Pseudomonas aeruginosa is a common opportunistic pathogen that can cause disease not only in plants and animals but also in humans. This ESKAPE Pathogen is of utmost importance because it is a multidrug-resistant pathogen, with very advanced antibiotic resistance mechanism, that can survive under various environmental conditions. According to the Centers for Disease Control and Prevention (CDC), it is the most common disease-causing species. P. aeruginosa affects different sites within the body, including urinary tract, skin (burn or surgical wounds), and the respiratory tract and causes severe acute and chronic infections in immunocompromised patients with cancer and patients suffering from severe burns and cystic fibrosis. It is often associated with hospital-acquired infections because there is a higher risk for infection if you have surgical wounds or burns or if you are being treated with a mechanical ventilator and other medical devices such as urinary or intravenous catheters.
General Cellular and Morphological Characteristics of the Organism
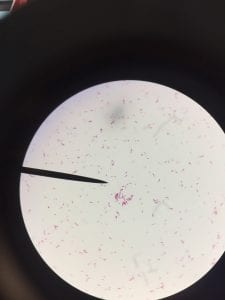
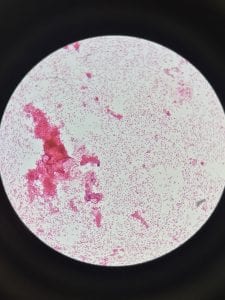
Pseudomonas aeruginosa is a Gram-negative, rod-shaped, asporogenous, and monoflagellated bacterium (Wu, 2014). Its size is about 0.5 to 0.8 µm, and it belongs to the bacterial family Pseudomonadaceae (Putty, 2007).
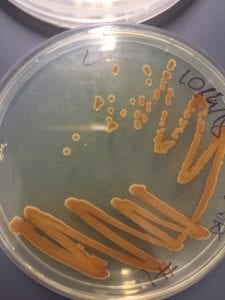
Pseudomonas aeruginosa is often identified by its pearlescent appearance and grape-like or tortilla-like odor (Putty, 2007). P. aeruginosa strains can produce one or more pigments: pyocyanin (blue-green), pyoverdine (yellow-green and fluorescent), and pyorubin (red-brown) (Wu, 2014).
Pseudomonas aeruginosa has simple nutritional requirements as it is often observed to grow in “distilled water” (Putty, 2007). It grows well at 25°C to 37°C, but it is its ability to grow at 42°C that help us differentiate it from many other Pseudomonas species (Wu, 2014).
Like most environmental bacteria, P. aerugionosa lives predominantly in slime-enclosed biofilms adherent to available surface from which it periodically releases (Putty, 2007). It is present in soil and aquatic environments. However, as mentioned before, it is tolerant of a variety of environmental conditions. It is also capable of growing in diesel and jet fuel, where it is known as a hydrocarbon utilizing microorganism (or “HUM bug”), causing microbial corrosion (Putty, 2007). Furthermore, it is resistant to high concentrations of salts and dyes, weak antiseptics, and many antibiotics (Putty, 2007).
Clinical Importance and Prevalence
The most difficult challenge when facing P. aeruginosa is its ability to rapidly develop resistance to multiple classes of antibiotics during treating an infection, ability to survive on minimal nutritional requirements and ability to tolerate a variety of physical conditions. These contribute to the organism’s capacity of persisting in hospital settings.
Data collected by the CDC National Nosocomial Infections Surveillance System from 1986 to 1998 reveals that P. aeruginosa was the fifth most frequently isolated nosocomial pathogen, accounting for 9% of all hospital-acquired infections in the United States; the second leading cause of nosocomial pneumonia (14 to 16%); third most common cause of urinary tract infections (7 to 11%); fourth most frequently isolated pathogen in surgical site infections (8%), and seventh leading contributor to bloodstream infections (2 to 6%) (Lister et al., 2009). In addition, more recent studies continue to show it is the leading cause among pediatric patients in the intensive care unit (Lister et al., 2009).
Infection
In hospital settings, infections by P. aeruginosa can be transmitted in hospitals by nursing staff, medical equipment, sinks, disinfectants, and food (Lister et al., 2009). Transmission occurs from improper hygiene, by patient contact with a contaminated reservoir or by ingestion of contaminated materials.
As described before, P. aeruginosa affects different sites within the body, including urinary tract, skin (burn or surgical wounds), and the respiratory tract and causes severe acute and chronic infections in immunocompromised patients with cancer and patients suffering from severe burns and cystic fibrosis (Wu, 2014).
Furthermore, exposure to contaminated water can also cause mild P. aeruginosa infections. For example, inadequately chlorinated hot tubs and swimming pools can cause ear infections and skin rashes. P. aeruginosa can also cause eye infections in users of contact lenses (Bennington-Castro, 2015).
Pathology
Bloodstream infections can cause:
• Fever and chills
• Body aches
• Light-headedness
• Rapid pulse and breathing
• Nausea and vomiting
• Diarrhea
• Decreased urination
Pneumonia can cause:
• Fever and chills
• Difficulty breathing
• Cough, sometimes with yellow, green, or bloody mucus
Urinary tract infections can cause:
• Strong urge to urinate frequently
• Painful urination
• Unpleasant odor in urine
• Cloudy or bloody urine
Wound infections can cause:
• Inflamed wound site
• Fluid leakage from wound
Ear infections can cause:
• Ear pain
• Hearing loss
• Dizziness and disorientation
(Bennington-Castro, 2015)
Ineffective Antibiotics
P. aeruginosa currently shows resistance to the following antibiotics: penicillin G; aminopenicillin, including those combined with beta-lactamase inhibitors; first and second generation cephalosporins; piperacillin; piperacillin and tazobactam; cefepime; ceftazidime; aminoglycosides; the quinolones; the carbapenems; colistin and fosfomycin (Hancock, 2000).
Effective Antibiotics
P. aeruginosa is most susceptible to the following antibiotics: cefepime, amikacin, ceftazidime, tobramycin, the combination of piperacillin and tazobactam, meropenem, imipenem, piperacillin, ciprofloxacin, gentamicin, and fosfomycin (Yayan et al., 2015).
Corresponding Safe Relative
The corresponding relative safe to P. aeruginosa is Pseudomonas putida. P. putida is a rod-shaped, flagellated, gram-negative bacterium that is found in most soil and water habitats where there is oxygen. It grows optimally at 25-30°C and can be easily isolated. Unlike P. aeruginosa, P. putida has a nonpathogenic nature; therefore, researchers find P. putida beneficial to research as it also happens to be very versatile and easy to handle (Marcus, 2003). For example, as P. putida assists in promoting plant development, researchers use it in bioengineering research to develop biopesticides and to the improve plant health (Espinosa-Urgel, 2000).
Works Cited
Bennington-Castro, Joseph. “What Is Pseudomonas Aeruginosa?” Stroke Center, Everyday Health, 7 Aug. 2015.
Espinosa-Urgel, Manuel, and Amparo SalidoJuan-Luis Ramos. “Genetic Analysis of Functions Involved in Adhesion of Pseudomonas Putida to Seeds.” Journal of Bacteriology, American Society for Microbiology Journals, 1 May 2000.
Hancock, R E, and D P Speert. “Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Impact on Treatment.” Current Neurology and Neuroscience Reports., U.S. National Library of Medicine, Aug. 2000.
Lister, Philip D., et al. “Antibacterial-Resistant Pseudomonas Aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms.” Current Neurology and Neuroscience Reports., U.S. National Library of Medicine, Oct. 2009.
Marcus, Adam. “Versatile Soil-Dwelling Microbe Is Mapped.” GNN – Genome News Network, 10 Jan. 2003.
Putty, Murali. “Pseudomonas Aeruginosa.” EMLab P&K, Mar. 2007.
Wu, Weihui, et al. “Pseudomonas Aeruginosa.” Molecular Medical Microbiology, Academic Press, 29 Sept. 2014.
Yayan, Josef, et al. “Antibiotic Resistance of Pseudomonas Aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period.” PLOS ONE, Public Library of Science, 2 Oct. 2015.