Assigned ESKAPE Pathogen: Enterococcus faecium
Why is this ESKAPE Pathogen of interest:
Enterococcus faeciumis normally found in human and other mammal gastrointestinal tracts as part of the gut microbiota. However, when located elsewhere, it can cause infections. This has become a large problem in hospitals, causing about 3% of sepsis cases in NICUs4. The emergence of VRE, or vancomycin resistant enterococci, in the early 2000s made this infection a larger issue, further compounding the growing problem of antibiotic resistance worldwide4.
General Cellular and Morphological Characteristics of the Organism (taxonomic classification, nutrition, cell shape, habitat):
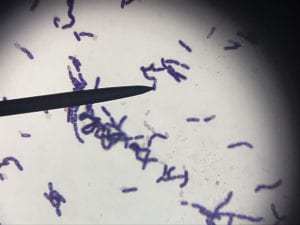
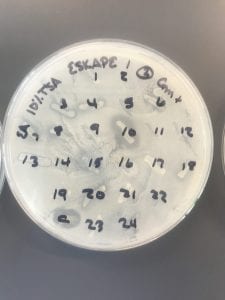
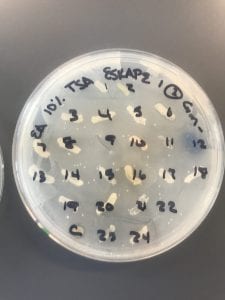
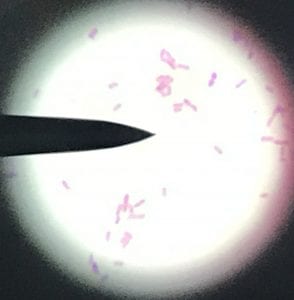
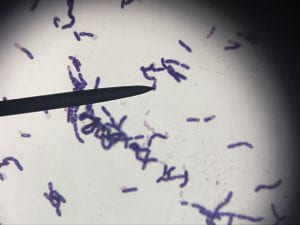
This is a gram-positive aerobic bacterium that grows best on Columbia blood agar at thirty-seven degrees Celsius for twenty-four to forty-eight hours1. It typically grow as diplococci, meaning they grow in pairs of round colonies, or as short chains of round colonies and will exhibit hemolysis on some blood medias8.
As previously mentioned, they typically live in the intestines. The Enterococcus genus used to be classified as group D of the Streptococcus genus; however, genetic analysis proved too large a difference between Streptococci and Enterococci to classify them in the same genus8.
Clinical Importance and Prevalence:
Nosocomial infections are a growing problem given both antibiotic resistance and the aging of the Baby Boomer population causing an influx of ill, elderly patients in hospitals. This organism is usually resistant to penicillin (about 90% of cases in one Australian hospital) and resistance to vancomycin has hit about 50% worldwide3. Over twenty-eight percent of people with these infections in the same hospital died within 30 days3. Some research has shown that fluoroquinolone use against these infections should be reduced, as it may play a role in the development of linezolid resistance6.
Infection:
E. faecium, along with its close relative Enterococcus faecalis, cause endocarditis, urinary tract infections, prostatitis, intra-abdominal infection, and cellulitis5. These infections typically occur from lack of correct hygiene methods in hospitals, such as handwashing4.
Pathology:
Symptoms vary based on the location of the infection:
- Endocarditis: fever, chills, fatigue
- UTI: pain during urination, foul smelling and/or dark/cloudy urine, polyuria
- Prostatitis: pain during urination, excessive night urination, urinary retention
- Intra-abdominal infection: fever, chills, fatigue
- Cellulitis: red skin, swelling, tenderness to the touch
Ineffective Antibiotics:
Strains isolated from humans have been shown to be resistant to vancomycin and linezolid (see above referenced sources). There is also evidence of strains resistant to chloramphenicol, tetracycline, ciprofloxacin, and erythromycin isolated from pigs in Malaysia5.
Effective Antibiotics:
The current recommendations include streptogramins, oxazolidinones, and Daptomycin5,6.
Corresponding Safe Relative:
The safe relative for E. faeciumis Enterococcus raffinosus. This organism has also been found to be vancomycin resistant and can cause endocarditis2. However, these infections are rare and the human immune system is normally able to withstand infection. Since the safe relative is in the same genus, it behaves similarly, but is less commonly the cause of infection in humans and less commonly resistant to antibiotics.
Sources:
- Culture Collections. Public Health England. Accessed October 5 2018.
- Dalal, Aman MD,Urban, Carl PhD, Rubin, David MD, Ahluwalia, Maneesha MD. Vancomycin-Resistant Enterococcus raffinosus Endocarditis: A Case Report and Review of Literature. Infectious Diseases in Clinical Practice. May 2008.
- Kelvin W. C. Leong, Louise A. Cooley, Tara L. Anderson, Sanjay S. Gautam, BelindaMcEwan, Anne Wells, Fiona Wilson, Lucy Hughson, & Ronan F. O’Toole. Emergence of Vancomycin-Resistant Enterococcus faeciumat an Australian Hospital: A Whole Genome Sequencing Analysis.Scientific Reportsvolume 8, Article number: 6274 (2018).
- Lakshmi Srinivasan, Jacquelyn R. Evans, in Avery’s Diseases of the Newborn (Tenth Edition), 2018
- Larry M. Bush, MD, Charles E. Schmidt, and Maria T. Perez, MD. Enterococcal Infections. Merck Manual. Accessed Oct 5 2018.
- Shiang ChietTan, Chun Wie Chong, Cindy Shuan Ju Teh, Peck Toung Ooi, Kwai Lin Occurrence of virulent multidrug-resistant Enterococcus faecalisand Enterococcus faecium in the pigs, farmers and farm environments in Malaysia. PubMed. August 2018.
- Thomas E. Dobbs, Mukesh Patel, Ken B. Waites, Stephen A. Moser, Alan M. Stamm, Craig J. Hoesley. Nosocomial Spread of Enterococcus faeciumResistant to Vancomycin and Linezolid in a Tertiary Care Medical Center. Journal of Clinical Microbiology. June 2018.
- Enterococcus. Accessed Oct 5 2018.