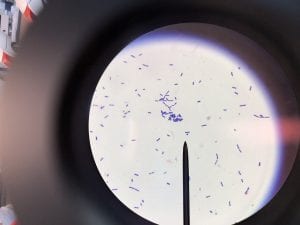
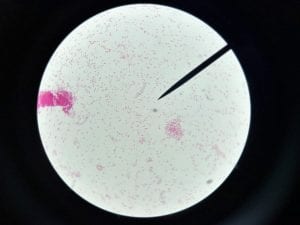
Acinetobacter baumannii is a multi-drug resistant, pathogenic bacteria that is commonly found in hospital settings. Being that is an emerging source of nosocomial infection, and is becoming more prevalent, Acinetobacter baumannii is of interest to the medical and scientific communities (1).
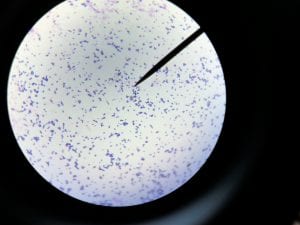
A. baumannii is gram negative, aerobic, and coccobacillus in shape (Figure 1) (3). The genus Acinectobacter was first discovered in 1911, when a Dutch scientist isolated microorganisms from soil. Originally “Acinectobacter” was used to designate non-motile bacteria. However, today it is currently the genus for “gram-negative, strictly aerobic, nonfermenting, nonfastidious, nonmotile, catalase-positive, oxidase-negative bacteria with a DNA G+C content of 39% to 47%” (2). Differences in the carbapenemase and gyrB genes of A. Baumannii distinguish it from its close relatives. A. baumannii’s “safe” relative is A. baylyi (2). A. baumannii, Acinetobacter genomic species 3, and Acinetobacter genomic species 13TU are extremely similar, and cannot be distinguished from one another using commercial methods. Therefore, the 3 species are often lumped together as a complex, and treated as one (2). Acinectobacters are commonly found in soil and water in nature, and some are part of the flora of human skin. However, virtually no individuals have tested positively for A. baumannii outside of hospital settings and combat zones, so the natural habitat of A. Baumannii is currently unknown (2). Very few individuals in China and tropical Australia have been tested positively for A. baumannii (5). In the laboratory setting, A. Baumannii grows well on Luria-Bertani broth (LB) (1).
As mentioned prior, A. baumannii is rarely found outside of the hospital setting. One notable exception to this is the prevalence of A. baumannii in skin infections among soldiers and other military personnel (1). Additionally, A. baumannii is well documented in burn units and ICUs. Immunocompromised individuals, as well as individuals with prolonged hospital stays are at the greatest risk for infection of A. baumannii (1). Hospital-acquired pneumonia and bloodstream infections, as well as post-neurosurgical meningitis, are the diseases commonly associated with A. baumannii. In 2003 the CDC reported that 7% of ICU-acquired pneumonias were caused by A. baumannii, and between 5 and 10% of cases of ICU-acquired pneumonia were due to A. baumannii (2). It is hypothesized that soldiers returning from Afghanistan and Iraq brought this pathogen into U.S. hospitals. A. baumannii has thrived in this setting due to its antibiotic and antiseptic resistance (2). Infections typically begin when an external device, such as a ventilator, is used in the treatment of a patient. The external device provides an entry site for A. baumannii, where it can then colonize (and can form biofilms)—for this example, in the lungs (1). Figure 2 below shows how A. baumannii spreads in the hospital setting, and Figure 3 shows how it affects the human body (6). Symptoms of an infection caused by A. baumannii are hard to distinguish among already ill individuals, however, they typically include fever, chills, and cough (5).
A. baumannii is known to be resistant to cephalosporins (can degrade beta-lactam), aminoglycosides, quinolones and, lately, carbapenems (3). When A. baumannii was first treated in the 1990’s, carbapenems were effective antibiotics, but are becoming less effective over time (3). Modified efflux pumps, porins, and lactamases enable A. baumannii to be resistant to several classes of drugs (4). The World Health Organization reports that A. baumannii is susceptible to polymyxins, but academic research has focused on new developments to combat gram-negative bacteria. Outer membrane proteins of gram-negative bacteria are being targeted experimentally as potential antibiotics (3). Additionally, bacteriophages are also being investigated as potential antibiotics, due to their ability to selectively target bacteria (1).
ESKAPE Pathogen Fig 1-27hgnn4
ESKAPE Pathogen Fig 2-18eokb8
ESKAPE Pathogen Fig 3-1t4ue2e
References:
(1) Howard, Aoife, et al. “Acinetobacter Baumannii.” Virulence, vol. 3, no. 3, 2012, pp. 243–250., doi:10.4161/viru.19700.
(2) Peleg, A. Y., et al. “Acinetobacter Baumannii: Emergence of a Successful Pathogen.” Clinical Microbiology Reviews, vol. 21, no. 3, 2008, pp. 538–582., doi:10.1128/cmr.00058-07.
(3) Soojhawon, Iswarduth, et al. “Discovery of Novel Inhibitors of Multidrug-Resistant Acinetobacter Baumannii.” Bioorganic & Medicinal Chemistry, vol. 25, no. 20, 2017, pp. 5477–5482., doi:10.1016/j.bmc.2017.08.014.
(4) Vila, Jordi, et al. “Porins, Efflux Pumps and Multidrug Resistance in Acinetobacter Baumannii.” Journal of Antimicrobial Chemotherapy, vol. 59, no. 6, 2007, pp. 1210–1215., doi:10.1093/jac/dkl509
(5) “WPRO | Multidrug-Resistant Acinetobacter Baumannii (MDRAB).” World Health Organization, World Health Organization, 7 July 2017, www.wpro.who.int/mediacentre/factsheets/fs_20101102/en/.
(6) Dijkshoorn, Lenie, et al., “An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii”, Nature Reviews Microbiology, vol. 5, 2007, pp. 939-951
https://doi.org/10.1038/nrmicro1789