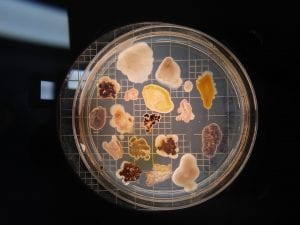
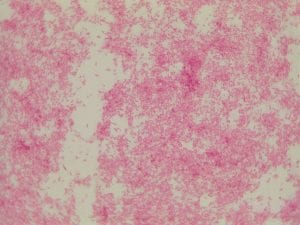
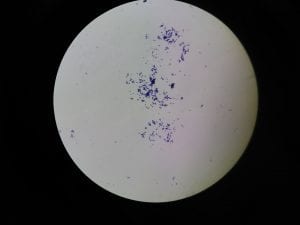
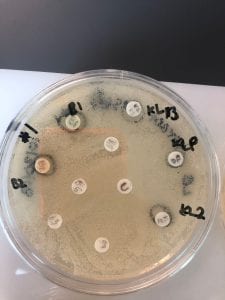
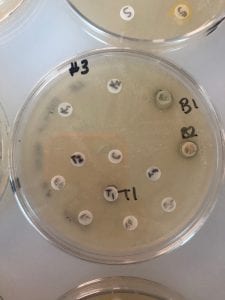
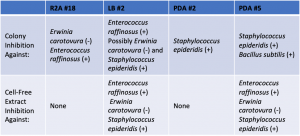
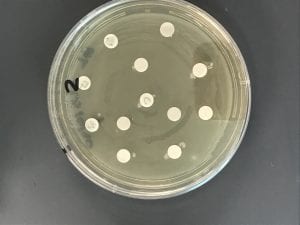
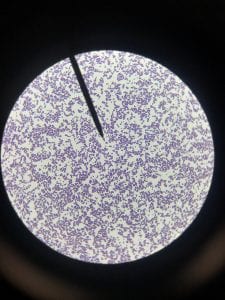
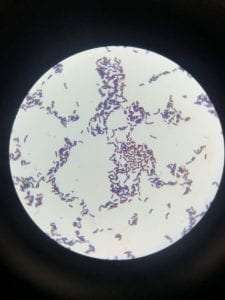
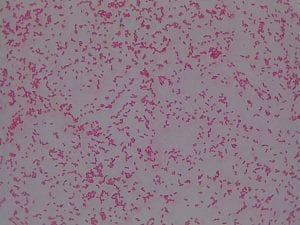
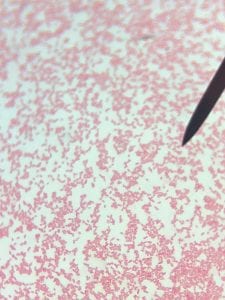
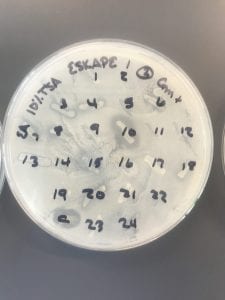
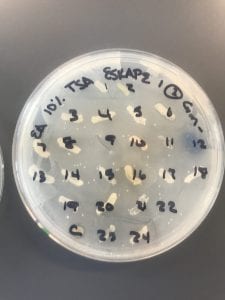
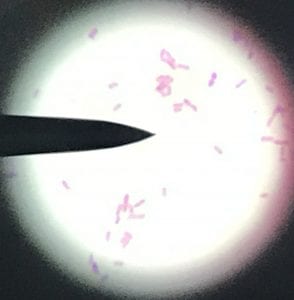
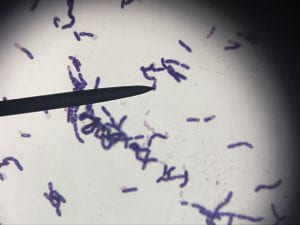
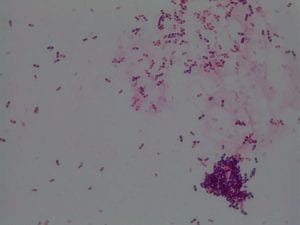
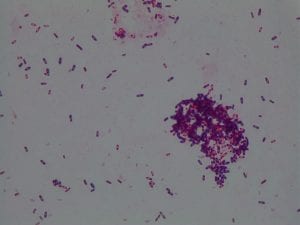
Assigned ESKAPE Pathogen
Enterococcus faecium
Why is this ESKAPE Pathogen of interest (in brief)
Enterococcus faecium can exist commensally within human or animal gastrointestinal tracts but can also be pathogenic in that it can cause neonatal meningitis and endocarditis (1). There have been strains of Enterococcus faecium identified as resistant to vancomycin – coined Vancomycin-Resistant E. faecium (VRE). Enterococcus faecium has also show tolerance to handwash alcohols in hospitals (2). Since alcohol-based disinfectants are used to control hospital infections worldwide, it is frightening that the multidrug-resistant Enterococcus faecium displays tolerance to commonly used hand rub alcohol solutions. VRE can survive on inanimate surfaces for weeks – including medical equipment – which explains how most VRE infections are acquired nosocomially (3).
General Cellular and Morphological Characteristics of the Organism (taxonomic classification, nutrition, cell shape, habitat)
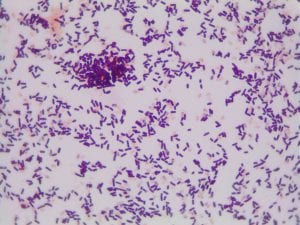
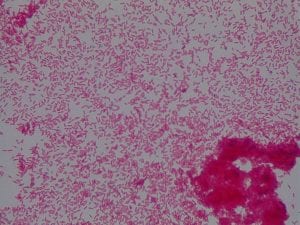
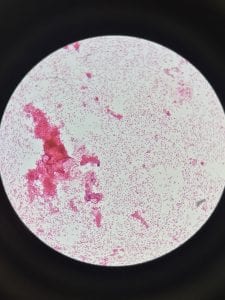
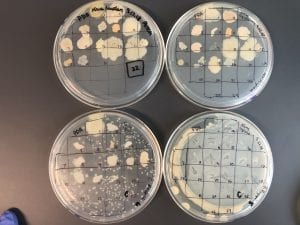
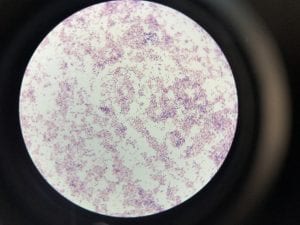
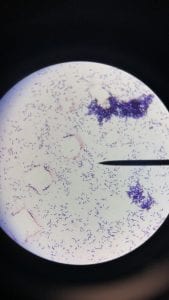
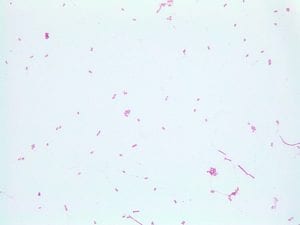
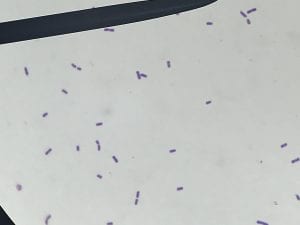
Enterococcus faecium belongs to Domain: Bacteria, Class: Bacilli, Order: Lactobacillales, Family: Enterococcaceae, Genus: Enterococcus and Species: faecium. This ESKAPE pathogen are Gram positive cocci (spherical in shape) that form short to medium length chains. They can also exist in pairs or single cells. E. faecium are also Facultative anaerobes that are ovoid in shape (1). They do not have cytochrome enzymes and are catalase negative (1). Enterococci are found in the feces of most healthy adults – where there are more faecalis than faecium although both are present. Lower percentages were found in oral cavities in healthy students (1). These bacilli tend to live in the gastrointestinal tract of humans and animals and live in feces and sewage. They are able to withstand harsh environmental conditions including high temperatures, periods of drying, and some antiseptics (1).
Clinical Importance and Prevalence
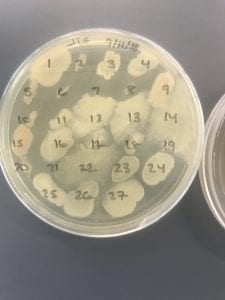
E. faecium’s high level of inherent and acquired resistance (especially VRE’s) along with the pathogen’s ability to survive on surfaces in hospitals makes it an important bacterium to study clinically. VRE constitute about 43% of all Enterococci isolates (4) making it a severe threat across the world, especially in hospitals that rely upon vancomycin or related antibiotics to combat infection. The bacterium has shown tolerance to hand-rub alcohols, making it a greater threat in hospitals (2). Additionally, this pathogen affects largely older adults with comorbidity, leading to heightened mortality rates (3). Furthermore, Enterococci harbor transferable genetic elements, meaning that resistant genes can be passed to both Gram positive and negative bacteria by conjugation systems with plasmids and transposons (4). This potential of E. faecium to pass on its multidrug resistance to other bacteria in a horizontal fashion is particularly frightening.
Infection (How does the infection occur and where is it localized?), Pathology (What disease is caused? What are the symptoms?)
E. faecium is known to cause urinary tract infections, intraabdominal, pelvic and wound infections, superinfections, and bacteremias (often with other organisms) (5). Lower urinary tract infections including cystitis, prostatitis, and epididymitis are seen in older men (5). Enterococci can also cause infection in the abdominal lining in conjunction with liver cirrhosis or in patients with chronic peritoneal dialysis (5). This ESKAPE pathogen is the third most common organism seen in nosocomial infections (3). Another notable infection of E. faecium is endocarditis. Bacteremia and endocarditis are common infections of Enterococci. The mortality associated with these infections is likely partly due to the demographic of patients who present: older adults with multiple underlying diseases such as diabetes. Synergistic, bactericidal attack is required for treatment of endocarditis (5). Multidrug resistant Enterococci are arising, including Vancomycin Resistant Enterococcus (VRE). Enterococcal surface proteins are virulence factors that contribute to infection in humans (5).
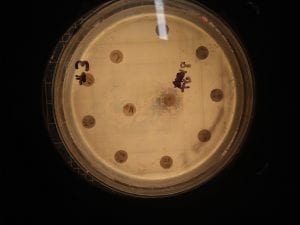
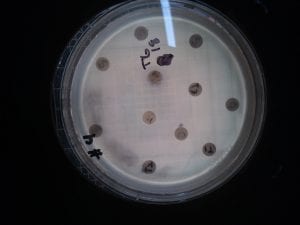
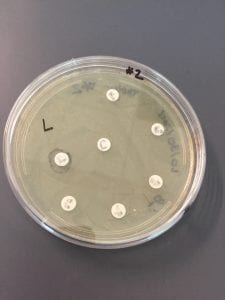
Ineffective Antibiotics (Antibiotics to which the organism has acquired resistance)
E. faecium has shown greater antibiotic-resistance than E. faecalis (3). More than half of its pathogenic isolates express resistance to vancomycin, ampicillin, and high levels of aminoglycosides (3). Additionally, the pathogen has shown resistance to some handwash alcohols (2). Enterococci also exhibit inherent and acquired resistance to cephalosporins, clindamycin, tetracycline, and penicillins. A mutation in the domain V of the 23 S rRNA of E. faecium appears responsible for linezolid resistance (5). Additionally, resistance to quinupristin-dalfopristin may be due to enzyme modification, and drug efflux (5).
Effective Antibiotics (Antibiotics known to inhibit the organism)
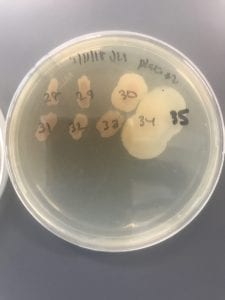
VRE can be successfully treated with sultamicillin (5). For most Enterococcal infections, single-drug therapies with penicillin, ampicillin, or vancomycin is adequate although resistance against these antibiotics has been observed (3). Currently, linezolid, daptomycin, tigecycline and the streptogramins (quinupristin/dalfopristin) have shown activity against VRE’s (3).
Corresponding Safe Relative
Enterococcus faecium’s safe relative is Enterococcus raffinosus (6). This non-faecialis and non-faecium enterococcus has rarely been connected to human infections (6). One of the first and only reported cases of E. raffinosus infection was endocarditis in an 85 year old man described in 2009 by Antonio Mastroianni in Le Infezioni in Medicina (6).
References
(1) Murray BE. (1990). The life and times of the Enterococcus. Clinical Microbiology Review. 3(1):46-65.
(2) Pidot SJ., Gao W., Buultjens A.H., Monk IR., Guerillot R., Carter GP., Lee JY., Lam, M., Grayson L., Ballard SA., Mahony AA., Grabsch EA., Kotsanas D., Korman TM., Coombs GW., Robinson JO., Silva A., Seemann T., Howden BP., Johnson PD., Stinear TP. (2018). Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Science Translational Medicine. 10(452).
(3) Dobbs TE., Patel M., Waites KB., Moser SA., Stamm AM., Hoesley CJ. (2006). Nosocomial Spread of Enterococcus faecium Resistant to Vancomycin and Linezolid in a Tertiary Care Medical Center. Journal of Clinical Microbiology.
(4) Olawale KO., Fadiora SO., Taiwo SS. (2011). Prevalence of Hospital-Acquired Enterococci Infections in Two Primary-Care Hospitals in Osogbo, Southwestern Nigeria. African Journal of Infectious Diseases. 5(2):40-46.
(5) Higuita NA. and Huycke MM. (2014). Enterococcal Disease, Epidemiology, and Implications for Treatment. Print.
(6) Mastroianni A. (2009). Enterococcus raffinosus endocarditis. First case and literature review. Le Infezioni in Medicina. 1:14-20.